This page is intended to be a starting place for students new(ish) to
R programming, specifically those in the UChicago course Data to
Manuscript in R. Many pieces of advice on this page are
generalizable to other programming languages; others are R-specific.
I’ve tried to keep things limited to minimally controversial ideas, but
you’re allowed to disagree with me here.
Whenever you read about best practices, there is really only one true
rule: The best code is the code that works.
Aiming to have code that is (1) standardized, (2) intelligible, (3)
maintainable, and (4) contextualized should help you create code that
works. On top of working, it should hopefully also help you operate in a
world of collaborative and open quantitative research. It should put you
in the best place for effective troubleshooting, for reproducing
analyses, and for returning to your code months or years from now. But
ultimately, these are guidelines that are intended to be
helpful. Try to incorporate these practices into your coding
workflow, but be patient and forgiving. If you have to choose between
code that’s pretty and code that works, choose code that works every
time.
Standardized
R is a pretty forgiving language. It’s whitespace insensitive (in
case you don’t already believe the tabs vs spaces debate is irrelevant)
and has relatively few forbidden characters or “arbitrary” syntax
constraints. If you break a rule (like you try to start a variable name
with a number) you get an error, which is a lot nicer than if it were to
just run anyway and produce mysterious, inexplicable results.
This has a lot of advantages, but one downside is that your code can
get extremely messy before things actually start to break. You can write
perfectly functional R code that is utterly indecipherable to humans,
including you-the-coder. Standardizing styling decisions helps keep
things organized and human-friendly.
Style guides
Coding style guides serve the same functions as publication style
guides, like AP Style or the New York Times manual of style. They demand
(or at least encourage) consistency, which should make things more
comprehensible for you and your “readers” (collaborators).
An effective and comprehensive style guide for R programming will
establish conventions for naming objects and functions, handling
whitespace and punctuation, and preferred syntax or representations
(e.g., a rule to use the <- assignment operator rather
than =). It will also likely contain guidelines for
otherwise highly subjective things like how to structure comments, how
verbose function definitions should be, how to clearly construct error
messages, and the necessary elements and formatting of
documentation.
The tidyverse style
guide is an excellent, comprehensive, and widely used style guide
for R. If you don’t already have a preferred style guide, I suggest
starting here.
Internal consistency
The best style guide is the one you can stick to. So long as (1) it
functions within your environment (no matter how much you’d like to
begin your variable names with numerals, it’s not a style option) and
(2) it’s comprehensible (for you totally and to others generally), the
internal consistency is what really matters.
Pay attention to how you manage to adhere to your style as you code.
Are there guidelines you can’t seem to remember or always mess up? Are
there things that just confuse you? Do you look back on your code that
perfectly follows your style and forget why you styled certain things
the way you did? These are most likely to be the breaking points for
internal consistency. If some aspect of style isn’t helping your code be
more comprehensible and more efficient to review and
maintain, it’s counterproductive and you should change to something that
will be more helpful.
For example, the tidyverse style guide recommends using underscores
when naming all objects. I find that using underscores for everything
leads me to mix up different kinds of objects. Is
child_gesture the name of the dataframe with gesture-level
coding or the column in that dataframe that contains frequency of
children’s gestures? Is shrug_bar_plot the modified
dataframe that is piped into a ggplot object, the ggplot object itself,
the chunk name where the plot is defined, the chunk where it’s rendered,
or something else?
It would be great if I was good enough at giving objects informative
names (see below!) to not confuse myself with this style, but
realistically it doesn’t work for me. Instead of using underscore
separators for everything, I opt to use periods for “complex” objects
dataframes, underscores for “simple” or “internal” objects like
variables, camel-case for “publishable” objects like plots, and hyphens
for “containers” like chunk and file names. Some people would find my
system infuriating for any number of reasons, but over time I’ve found
it works for me. Since I can be consistent with it, it really speeds up
coding and debugging, and it doesn’t actively impede comprehensibility
for others. That’s ultimately what matters.
Final notes on styling
While standarizing your coding style is an essential “best practice,”
remember that in the end styling is usually not make-or-break for your
code. I do encourage you to aim for an internally consistent style, but
don’t get lost in the weeds. Again: The best code is the code
that works. You have limited time and energy to work on your
project. Before you get too deep into developing your own personalized
style or rigorously checking every detail of every script you’ve ever
written to retroactively enforce a style, think about whether that’s
truly the most productive use of your precious time. Hopefully at some
point style really can be high priority, but if that’s not where you’re
at right now, that’s fine!
C Programming Style Guide Cartoon – “I hate
programmers.”
And one final recommendation: choose where to be cautious. It’s easy
to avoid special characters even if they’re technically permissible
sometimes, so just avoid them. Being concise is nice, but it’s probably
safer to prioritize readability over conciseness. Keep your future self
in mind. What can you do now to best help future-you when you come back
to this project in a year having completely forgotten everything you’re
doing now?
Intelligible
Meaningful naming
For some reason every example in every programming tutorial ever will
teach you how to create a variable with the name
my_variable and a function with the name
my_function. I get it, but these are objectively horrible
names. Imagine looking through someone’s code and seeing
my_function(my_variable, 2). What?? That better come with
like 12 lines of comments because otherwise it’s total nonsense.
Of course, no one actually recommends using my_whatever
in your actual code, but it’s the principle of the thing! Designing
object, function, and file names that actually tell you any useful
information can be a lot harder than it sounds, but it’s a game-changer
to the intelligibility of your code.
“There are only two hard things in Computer Science: cache
invalidation and naming things.”
— Phil Karlton
The big rule here (vs a guideline): Names should
describe the named thing.

“Keep it simple, stupid – commitstrip.com
A few guidelines for crafting meaningful names (source):
- Avoid disinformation. Don’t include
_df in the name of a data.table, don’t name a
function get_number if it returns a string.
- Use pronounceable names. Whether you want to or not
you’ll inevitably talk about your code out loud (even if just to a rubber duck). It’s a lot
easier to ask someone to check for errors in your
child_gesture table or your group_SES_quartile
function than cgtbl or grpsq. This doesn’t
mean you can’t abbreviate: chi_gest and SES4
are perfectly pronounceable too (though less transparent, which is a
trade-off you’ll need to consider).
- Use searchable names.
x is a useful
variable in math, not in programming. Plan ahead for when you need to
cmd+f to replace all cases of a name with a slightly different version.
Examples will often use p to name plots, but replacing all
ps with pcgChildScatter will leave you with a
lot of pcgChildScatterivot_longer()s and
ungroupcgChildScatter() and
pcgChildScattercg.child.compcgChildScatterares.
- Pick one word/format per concept. You turned a
sprawling data frame into 3 manageable intermediate data frames based on
groups of related measurements. Call them
gesture.freqs,
gesture.rates, and gesture.ratios rather than
count.gestures, summ.rates.gestures, and
grats.
- Avoid encodings. this is a little more complicated
(look at the source link above for more), but the gist is that names
should make sense on their own and not implicitly rely on knowing what
other things in the code do. The purpose of the function
mean_cgfq may be decipherable if you know that the
dataframe child.gesture exists and contains a
frequency variable, but out of context it’s a mystery.
Documentation
Documentation doesn’t usually need to be a major concern for beginner
or intermediate programmers. However, you’ll be depending on a
lot of documentation even if you’re not creating it yourself. As you
start wading through the world of R packages, pay attention to the
differences in documentation. Packages published to CRAN must adhere to
minimum standards of documentation. Many larger packages will have their
own websites or github repos (in addition to CRAN) with extensive
documentation, examples, and FAQs. Smaller independent and specialized
packages might have extremely useful functions but leave you desperately
trying to work out how to use them effectively.
Any time you construct something usable – anything from a simple
in-line function to a package to a shiny app – practice coherent
documentation. Does your documentation look like the kind of
documentation you find helpful? If you hand it off to a friend without
any additional guidance can they figure out what to do with it?
Think of multi-line comment chunks as a form of mini-documentation.
Explain what the point of a script is at the top of a file, explain what
the point of the function you’re defining is and what parameters it
accepts just above the definition, etc.
Maintainable
Aim to create code that is future-proof and collaborator-proof
(especially bad-collaborator-proof). It can be hard to know in
the moment whether you’re writing a script you’ll never open again or
one you’ll return to for years to come. Set yourself up for the
latter.
Lifecycles
R is constantly evolving, and R functions exist in “lifecycles.”
In general, you want to prioritize using functions that exist in the
stable stage. A stable function is currently maintained, works
with the most current version of R (and any dependent packages), and
does not currently have a better option.
Note the use of “current” in those descriptors, though. Any or all of
those may change, and it’s important to keep on top of things in any
code you hope to come back to in the future.
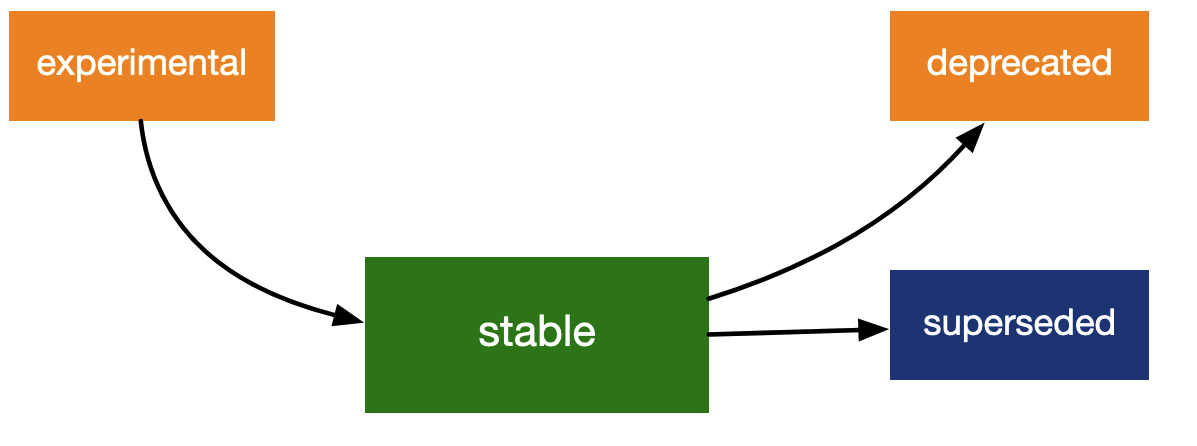
Lifecycle stages - sourced from
lifecycle.r-lib.org
/
R function lifecycle stages include:
- Experimental. This function is in development. Like
beta software or an early access video game, this function may or may
not be developed into a full, stable release. It may end up replacing
currently stable functions, or it may be abandoned tomorrow.
Experimental functions can be very helpful for very specific use-cases
where stable functions don’t yet exist or aren’t sufficient, but you
should use them with caution and add a comment noting that it’s
experimental in case it breaks your code when something updates.
- Stable. This function is currently operational,
up-to-date, and maintained. This is the default stage, so unless the
documentation indicates otherwise you can presume a function is stable.
You should prioritize functions in the stable life stage.
- Superseded. This function is still supported and
isn’t going away any time soon, but it’s not going to get any further
development. It is safe to use, but there is now an alternative
stable function that is preferred for its use. A good example
here is the
gather and spread functions which
have been superseded by pivot_longer and
pivot_wider. Gather and spread were around for a long time
and are essential to a lot of operational code, and a lot of people are
used to them now and don’t really want to switch over to something new;
it would be impractical to not support them. But the pivot options are
just, well, better. If you have a good reason to favor a superseded
function, go for it. If you’re not already familiar with it though, it’s
wiser to spend your time learning the newer, stable replacement.
- Deprecated. This function works now but won’t for
long. It may already not work with the current version of R, but still
works with some operational older versions. Only use deprecated
functions as a temporary last resort, and update with stable or
experimental functions once you’re able.
Learn more
about lifecycles here.
Data lifecycle
Your data don’t exist in quite such a formal “lifecycle,” but you can
thing about them in a similar way as you try to future-proof your
code:
- How much could your data change?
- Incomplete datasets will get more data (e.g., pilot datasets ->
final datasets)
- “Complete” datasets may eliminate some data (e.g., retroactively
excluding participants or measurement timepoints)
- Variables may need to be combined, anonymized, mutated, etc.
- How similar are your data to other data?
- Follow-up studies and replications usually need to make minor (and
sometimes major) changes to data format/organization
- Problems in data collection stage that are too late to change will
mean new data format when they are fixed next time
- Other researchers in your area who could benefit from your code
might use similar data collection methods but different organization
methods
DRY programming
“DRY”
programming stands for “Don’t Repeat Yourself.” Generally, stop
copying and pasting! There are certainly merits to copy/paste when
you’re making use of external code (e.g., from stackoverflow,
chatGPT suggested code, forking a repo, your own old project), but DRY
coding means minimizing repetition internally, within a single
script or project.
When you stay DRY, you:
- Avoid:
- Propogating mistakes that must be corrected individually
- Duplication conflicts
- Combing through verbose scripts for minor errors
- Promote:
- Replicable, reproducible code
- Abstracting code for multiple contexts of use
- Code usable by and relevant to other people and future-you
Abstracted coding
“Abstracting” in the sense of DRY coding usually refers to practices
like defining one function to use repeatedly rather than copying and
pasting the same block of code and changing variable names every time.
Abstraction exists at a finer-grained level as well, such as with paths
and variables. Generally, abstracted (relative) paths and variables are
more durable that hard-coded (absolute) ones.
Paths
Maximally abstracted file path references minimize broken links. As
an example:
Natalie's Work MacBook/Users/Natalie/repos/d2m/example-repo/images/barplot.jpg
- Only works on Natalie’s work computer no matter what
~/repos/d2m/example-repo/images/barplot.jpg
- Works on any machine if it’s cloned to this particular location
/images/barplot.jpg
- Works when you clone the repo anywhere as long as the internal
structure is unmodified
Variables
Your data will change. You want to set yourself up for making as few
changes as possible to accommodate changes to your data. This is the
same logic as writing your manuscript in R Markdown with in-text code
and references!
my_mean <- (2 + 4)/2
x <- 2 ; y <- 4�or
x <- 3 ;
y <- 10�the_mean <- (x + y)/2
- Output will change to reflect changes to 2 input variables
number_list <- c(2,3)�or
number_list <- 2:8�or
number_list <- c(1,2,8,100) ;
�a_mean <- sum(number_list)/length(number_list)- Output will change no matter how many input numerals are
averaged
Contextualized
Project priorities
Above all else (or I guess below, since this is the bottom of the
document, but I said this at the top too I think), the best code
is the code that works. You have limited time and resources.
You have needs and goals specific to your project and to you as an
individual. People will refer to best practices “rules,” but they are
called “best practices” for a reason. They are not hard and fast rules;
they aren’t supposed to be. They are what should be
relatively the most useful most of the time. Get in
the practice of using these guidelines when you can, but
ultimately this is not what your work is about.
Keep your project’s goals as your top priorities. Consider the
context of your work and accept that it’s not going to be exactly the
same as any other context, and it will be vastly different from many.
Make these guidelines work for you, stay flexible, and don’t worry if
and when you need to just say screw this and move on with your life.
Other
R Notesbooks and Markdown
Don’t use the visual editor!
Remember everything will run top to bottom, just like a regular .R
script. Right after your YAML header, your first chunks should 1) load
all libraries, 2) set document defaults (optional), 3) set a random seed
(optional), and 4) source any required external scripts in a sensible
order.
Code chunks
Code chunks should:
- Do 1 and only 1 thing. A chunk should serve a
single and transparent purpose. As a rule of thumb, if you can’t
describe what your chunk does in 5 or fewer words, it should probably be
more than one chunk. For example, one chunk might: assemble a ggplot,
render a plot or table, import or export intermediate datasets, filter
data for a specific purpose, print or store the output of a regression.
This means you’ll sometimes need multiple chunks for things that at
first feel like one big thing.
- Example: Include a demographics table in your document
- Chunk 1: Create an intermediate dataset with only relevant
demographic variables and observations
- Chunk 2: Perform any alternations or calculations (e.g., summarize
by gender groups, create income brackets)
- Chunk 3: Store a basic kable or table
- Chunk 4: Render the kable in the document with the aesthetic details
you want
- By separating it into multiple chunks, you can easily refer to
distinct elements. Maybe you want to create the same demo table for a
different dataset. Easy! You’ll have a new Chunk 1 but 2-4 are the same.
Want to call a value from the table in the text? It’s pretty messy to
pull that from the “pretty” kable you rendered, but it’s simple to pull
from the basic kable skeleton you built in chunk 3. Want to print a
complete demo table in one place and a summarized table somewhere else?
Make an alternative version of chunk 2 for the second table.
- Have informative and unique names. Name your chunks
to minimize human error. In addition to helping you stay organized, this
will help tremendously with troubleshooting. Error messages during the
knitting process don’t always give you the precise line number, but will
always give you the chunk name where the problem occurred. (This is also
a good reason to keep your chunks short and for 1 purpose only.) Aim to
have the chunk name clearly indicate the one “thing” your chunk does. By
default, I usually use verb-noun names:
read-data,
summarize-demos, build-child-table,
build-gesture-bar-plot. The only exception (for me) is
chunks that do the actual rendering of tables or plots, which I give a
simple label matched to the chunk that built it (e.g.,
build-gesture-stacked-bar saves a plot to the variable
gestureStackedBar which is then rendered in the chunk named
fig-gesture-stacked-bar).
- Critically, your chunk names should be:
- Unique (knitr will throw an error for duplicate chunk names)
- Informative (
fig-gesture-stacked-bar and not
bar-graph)
- Conventional (conservative and correct)
- e.g., R variables cannot start with a numeral, so even though chunk
names legally can, it’s better to avoid it
- Don’t use underscores or spaces. Even if they “work” they often
won’t work. Stick to
. and - as
separators.
- Specify whether/how they should run and print. Many
chunk options are available here, but at a basic level use
echo and include to tell knitr to show/hide
your code and/or output and use eval to say whether to run
the code at all. You can easily change these chunk settings in RStudio
by clicking the gear icon in the top right corner of any chunk.
- Show output only:
echo=FALSE
- Show code and output:
echo=TRUE
- Show nothing (run the code without printing anything):
include=TRUE
- Show nothing and do not run code (just ignore the whole chunk):
eval=FALSE, include=FALSE
- Be placed at the point they are needed or
referenced. In theory, you could place every single chunk at
the top of the file so long as they are in the correct order relative to
each other. In practice, that’s impossible to manage. The beauty of R
notebooks is the seamless integration of code and narrative, making the
whole thing as human-friendly as it is machine-friendly. Keep your
chunks as close as possible to the point in the narrative that they are
referenced to easily edit both narrative and code as you work.
- Be (relatively) short, sourcing longer scripts if
necessary. Using
source() will run the full code
of another file. Long blocks of code or instances where the purpose of
code really can’t follow the “one-chunk-one-thing” rule may call for
sourcing the code as a separate script. This is especially useful for:
- Code that you know you will rarely or never need to make dynamic
edits to, like a script that imports multiple .csv files, wrangles many
sources of data, and produces multiple intermediate datasets that will
be the essential data for your whole manuscript.
- Scripts that you run often that aren’t project specific. For
example, I have a generic
startup.R script I source that
loads my most commonly used libraries, sets my preferred ggplot theme
tweaks, and defines color palettes.
- Dedicated scripts for defining all the custom functions necessary
for the project. Nearly all of my projects source a
functions.R script, which all start from a basic file that
has functions I use in most projects (like one that calculates SD/SE and
adds error bars to plots).
GitHub & Collaboration
- Include and maintain both a .gitignore and README.md file in the top
level of your repository
- Make purposeful and wise decisions about managing public, private,
and protected data and files
- Use informative commit messages
- Pull before you start editing; commit as you work; push when you
close your session

Git xkcd
/
Do not forget that the whole point of using github is version
control! Do not create a new file for each assignment.
Your repo is the home of your research project, not a collection of
notes and homework. Keep it organized, future-proof,
collaborator-friendly, and contextualized beyond this
class.